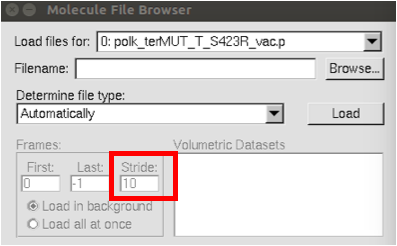
For the normal modes analysis, you need to have previously finished the cpptraj steps to create trajectory file for all the production steps. The topology file can be opened with vmd from the command line, but because the coordinate file is really large, it needs to be loaded in from the user interface. When loading in the data for the prmtop, set stride equal to 10 so that it only loads every 10 frames (see the image below).
$ vmd -prmtop vacuum.prmtop
Setting Stride
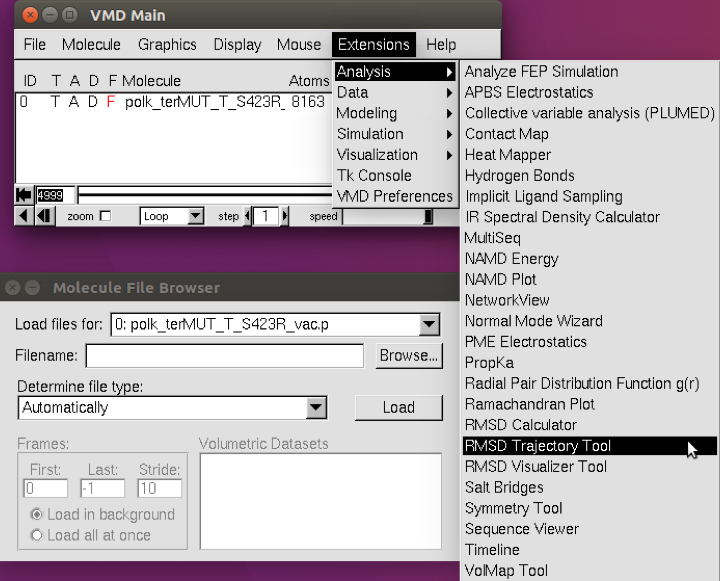
Once the trajectory files are loaded in, follow the VMD menu through
Extensions → Analysis → RMSD Trajectory Tool
(see below).
RMSD Analysis
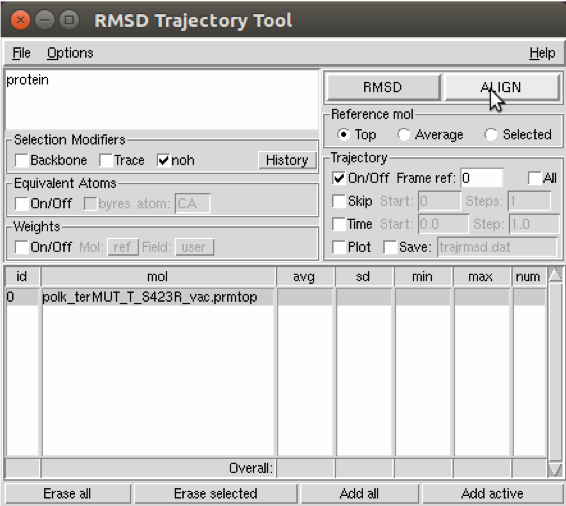
This brings up the menu (shown below) with several options. Click align.
RMSD Trajectory Tool
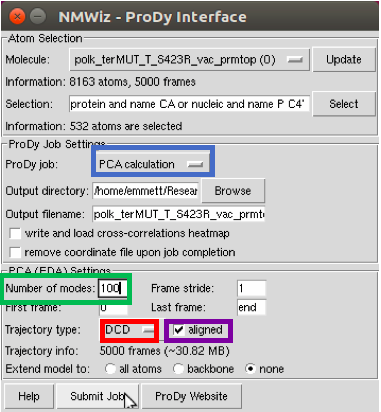
After the structure is aligned, proceed to the Normal Mode Wizard. It can be found in the same menu as the RMSD Trajectory Tool, shown previously.
From the NMWiz menu, select ProDy Interface. This pulls up the ProDy window. Several things need to be changed in this interface, including setting the ProDy job to a PCA (Principle Component Analysis) calculation, changing the number of modes to 100 (it can analyze every mode, but the only important ones are the first few), changing the trajectory type to DCD, and clicking the aligned structure option. The aligned structure option is what was performed using the RMSD Trajectory Tool, and saves time in the normal mode calculation. After changing each of these, the job is ready to be submitted.
ProDy Interface
Once submitted, a new NMWiz window appears (see below). The mode that is being visualized appears in the upper left. From this window (and several successive windows), you can save the information for the normal nodes for further analysis and comparison. To do this, select Plot Mobility.
NMWiz Window
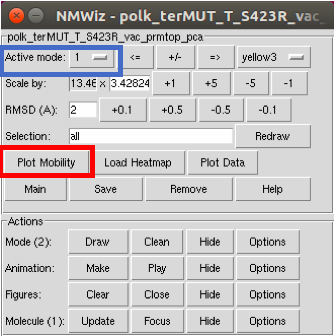
Plot Mobility brings up a plot (amazing!).
The data from this plot need to be saved in a roundabout way.
To start this process, go to File → Save as xmgrace.
Title this file however you want, but recognize this will be done for more than
1 mode. Once saved, the Grace window will appear.
In this window, follow Data → Export → ASCII.
Yet another Grace window will appear, allowing you to save the data as a .dat
file (see below).
To actually save, first title the new file (under the selection box), and then
click on the set in “Write Sets” that you want to save.
Then, click OK.
It won’t look like it did anything, so if you’re unsure if you clicked it,
click OK again and it will ask if you want to overwrite the file.
xmgrace Window
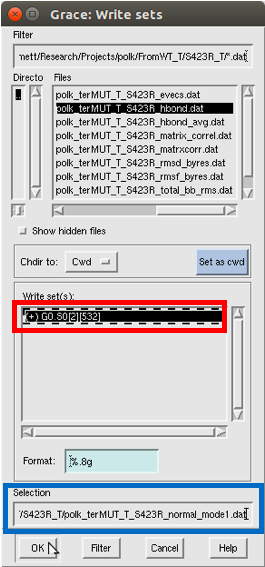
Follow this process for the first 3 normal modes (1, 2, and 3).