VMD has quite a few options for changing the view of your system.
These are found by following Graphics → Representations in
the VMD main menu. For any loaded system, the default (unless you’ve changed
some settings) is to show everything using the Lines drawing method.
Lines is helpful sometimes, but overwhelmingly, there are some common trends in
how people create images for publication/presentations, and those are typically
the representations that people use when visualizing their data all the time.
To start, type protein into the Selected Atoms box and press enter.
Then change the Drawing Method to New Cartoon and hit enter again.
Now you have created your first representation for the protein.
To add more layers, hit the Create Rep button.
VMD will automatically generate a second representation of what you just made,
and now you can edit that, hitting enter after every change.
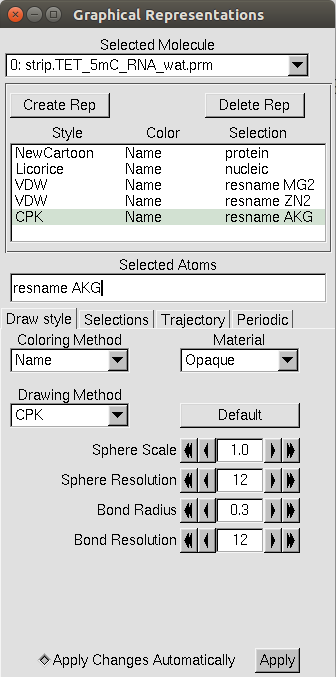
Other common trends for VMD visualization are to show metal ions using the
VDW drawing method (please… I’m begging you… call them “spheres” and
NOT “van der Waal’s balls”), using the Licorice drawing method for nucleic
acids, and showing any cofactors with the CPK drawing method.
However, you should really use what makes sense to you, until you’re told to
change it. The advice I have for you is to play around with these settings.
Two quick other notes: 1) under Materials, there’s a Transparent option,
which while it may not look it onscreen, will actually look transparent in an
image and 2) Coloring Method has a Color ID option, so that you can make
entire portions of your protein one color.
Finally, an example of a complete representation is shown below.
VMD’s Syntax
Ah, yes, you’ve just learned about Selected Atoms.
Now’s a good time to let you know VMD is a grammar-obsessed jerk who wants
everything to be stated in exactly the right way.
Here’s a list of things that VMD will accept:
protein: the proteinnucleic: any nucleic acid residuesall not water: everything in the structure that isn’t water. Any keyword, like nucleic or protein, can be used here.all not resname MG2: everything in the structure that doesn’t have the residue name MG2.resname MG2: anything in the PDB with the residue name of MG2. Any residue name (cough think of non-standard residues here cough) can be used with theresnamecommand, provided it’s found in the protein structure.resid 244: the residue corresponding to the number 244 in the PDB.all within 5 of resname MG2: everything in the structure within 5 Å residue name MG2.resid 1 to 125: all residue numbers from 1 through 125
To summarize: it’s powerful, but if you mess up, VMD won’t necessarily let you know that, and you’ll just think you’ve lost a critical part of your structure (like a zinc in your active site that they entirety of everything you’ve ever cared about in research). When in doubt, save a PDB and check it using gedit or vi for what you think is missing.
Saving/Loading Graphical Representations
If you’re going to be making a lot of images, or just revisiting the same
structure files over and over and over, you’ll probably want to save
visualization state.
This essentially saves the information for the loaded compound, such as frames
loaded in, and the graphical representation information.
To do this, either follow File → Save Visualization State
in the main menu, or type:
$ save_state name-of-saved-state.vmd
into the command line where VMD is operating from.
To open visualization states, either follow either follow
File → Load Visualization State in the main menu, or type:
$ vmd -e name-of-saved-state.vmd
when loading VMD from the command line.