MolProbity is a webserver that can be used to add hydrogen atoms to crystal structures, flip ASN, GLU, and HIS residues, and check the overall structure for bad contacts.
After determining the protonation states of titratable residues, upload the structure to MolProbity. The system will then be processed and remove all the hydrogens. The next screen is the selection menu.
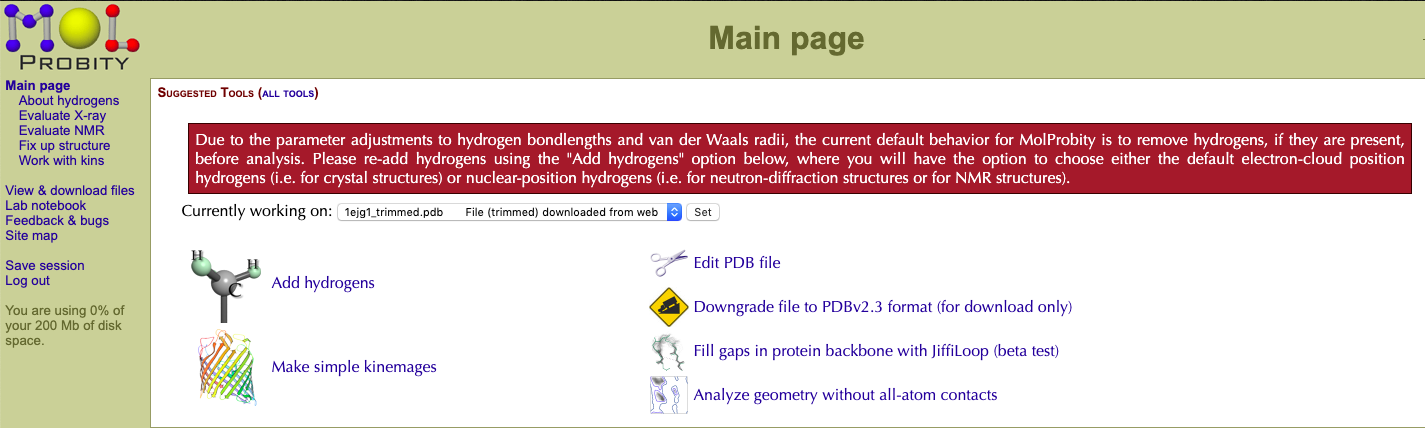
Select the Add hydrogens button to add the hydrogens back to the structure.
On the next screen, choose Asn/Gln/His flips (the advanced options don’t
matter).
For the x-H bond-length selection, choose the one that matches the PDB’s
experimental method.
As the description says, choose Electron-cloud x-H for x-ray crystal
structures and Nuclear x-H for NMR or neutron diffraction structures.
After a few more pages with details on adding hydrogens or flips, you’ll get to
a new selection menu. Now you’ll have the choice to Analyze all-atom contacts
and geometry.
Select this option, keeping the FH version of the PDB selected.
The default selections are likely fine, unless you need a particular setting.
The all-atom contact analysis gives a clashscore. Structures with less than 90% likely need to be fixed a little bit. The downloadable files from MolProbity will provide insight into which residues are clashing, and you can use a program like Chimera to select rotamers at those positions.