PDB stands for Protein Data Bank and is a file extension type that is used for crystal structures by the RCSB PDB. Once resolved, crystal structures are published on the website (see below), which provides information about the specifics of the crystals structure and relevant citations for the protein of interest. Published structures are assigned a PDBID, which is an identifying set of numbers and letters for that specific structure.
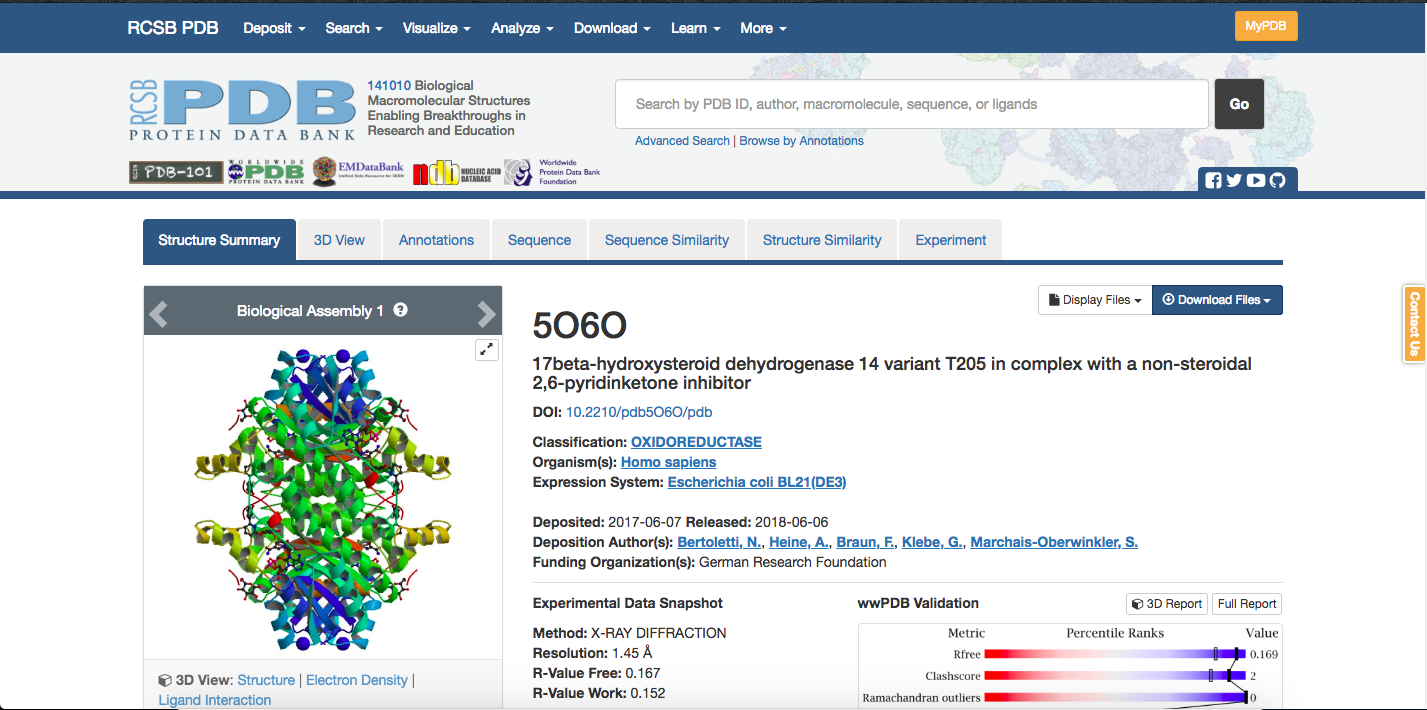
The PDB file (here being the data file with a .pdb extension) can be
downloaded from a box on the right-hand side of the structure’s page,
shown below.
This file will contain a lot of extraneous information,
which is why you will likely end up using a command like:
$ grep -e '^ATOM\|^HETATM\|^TER\|^END' PDBID.pdb > PDBID_clean.pdb
to extract out any lines that start with ATOM, HETATM, TER, or END, and save them to a new file.
The data in the PDB format is arranged into the following columns:
- Record Type
- Atom Number
- Residue Name / resname
- Chain Identifier (It is likely everything is the same chain)
- Residue Number / RESID
- X orthogonal coordinate
- Y orthonogonal coordinate
- Z orthogonal coordinate
- Occupancy
- Temperature Factor
- Segment ID (Essentially obsolete by everyone/thing except Chimera)
- Element Symbol
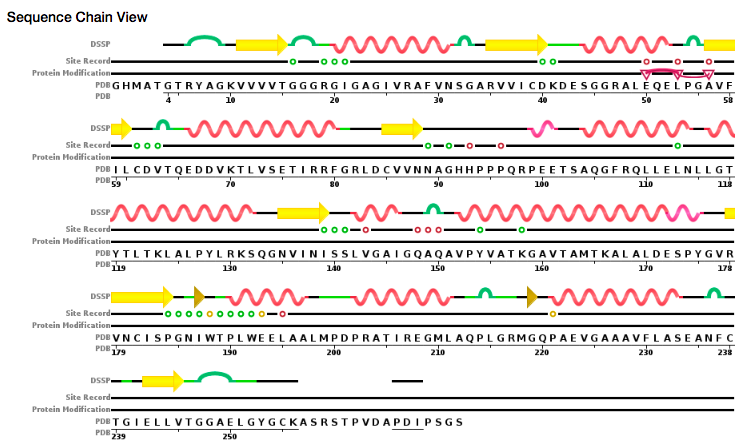
A very important part of the PDB page, especially when dealing with
SNPs is the sequence tab for a given
structure.
The information on that tab connects the computational structure with the
actual numbering of the protein. For instance, in the figure below,
the downloadable PDB file sets has residue 1 as G, but what
experimentalists consider residue 1 is actually an M.
Knowing this conversion is not only important so that the correct residues are
mutated for SNP studies, but also for working with collaborators and publishing
results.