A number of studies recently have focused on what single nucleotide polymorphisms (SNPs) do to a protein’s function. Changes in a single nucleotide that are found in less than 1% of human populations are SNPs. These SNPs can be missense mutations. PDB files generated from a crystal structure are typically in the wild type, meaning they do not contain the SNP. Changing a wild type to a known SNP can be performed using the program UCSF Chimera.
SNPs should be added after the wild type structure has been prepared (i.e. any linkers or specific modifications to match an experimental study have been added). After the wild type structure has been prepared (and saved), then in can be opened in Chimera. It would be wise to make a copy first, so that you do not accidentally overwrite the wild type structure.
Once the structure is pulled up, follow
Favorites → Command Line in the menu bar to pull up Chimera’s
command line.
In the command prompt, type the following command, where 123 is the residue
number of interest.
The specific number will be dependent on the protein numbering on the
Protein Data Bank website
discussed here.
select :123
This selects the specific SNP position, and should show it highlighted in
green. To then visualize the residue as a stick, follow
Actions → Atoms/Bonds → Show in the menu bar.
Using the menu bar again, follow
Tools → Structure Editing → Rotamers to bring up the rotamers
screen (see the image below).
The rotamers screen allows the current residue in the SNP position,
described as the rotamer type (THR in the rotamers image),
to the residue changed in the SNP.
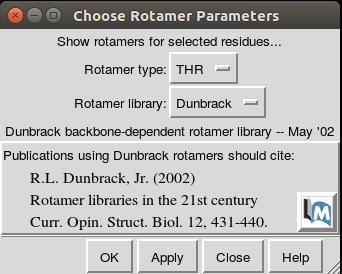
After changing the residue in the drop down menu, select OK.
You will then be given probabilities to pick from as to how likely it is that
that specific rotation of the amino acid would occur.
Clicking on each one will show the proposed orientation.
Most likely, though, the highest probability rotamer will be the best choice.
The “Existing side chain(s)” box should remain as replace,
as the residue is being replaced by the changed residue.
The probability that is highlighted is the one that is being used to replace t
he original residue, and its stated probability should be written down in your
lab notebook before changing it with OK.
Once the residue is changed, then a new PDB should be saved, indicating the SNP
in the file name. This can be achieved through File → Save PDB.
If multiple things are open in Chimera, highlight the appropriate structure
under Save models.
Specify the file location, hit save, and then do a happy dance;
you just mutated a residue in Chimera!