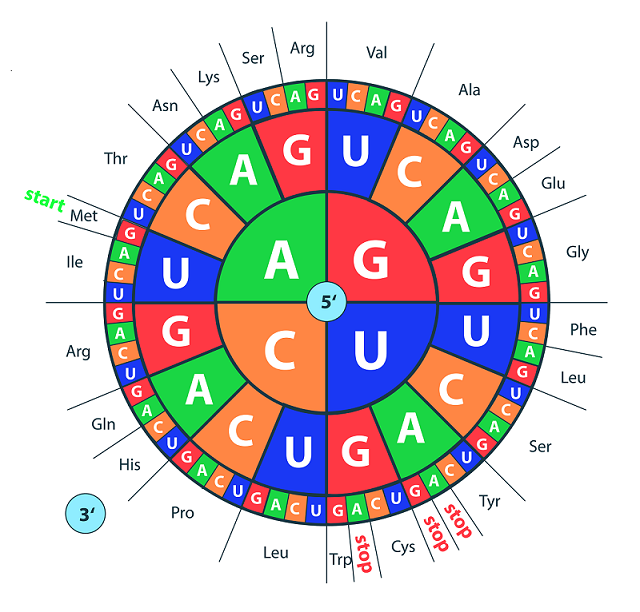
The central dogma of biology is “DNA makes RNA makes protein.” DNA consists of base pairs of adenine, cytosine, guanine, and thymine. These four bases (which change to adenine, cytosine, guanine, and uracil in RNA) end up becoming proteins through translation. In translation, three of these bases, known as codons on messenger RNA, code for a specific amino acid. Which amino acid, or residue, that they code for can be found by looking at the codon wheel (shown below). Sequences of these amino acids are what make up proteins.
Figure: Codon Wheel
Different mutations in DNA can propagate through to proteins. Some mutations, known as point-nonsense mutations, result when a mutation at a single residue results in an early stop codon. If a situation like that occurs in a protein that is critical for development, it is unlikely to result in a viable fetus. Similarly, frameshift mutations occur when the addition or loss of a DNA base messes with the reading frame of a group of 3 DNA bases. Thus, single or duplicate insertions would change the outcome of the protein, usually rendering it nonfunctional. Missense mutations are point mutations where a single nucleotide change results in a different amino acid. Other types include insertions, deletions, duplications, and repeat expansions, which will not be discussed here.
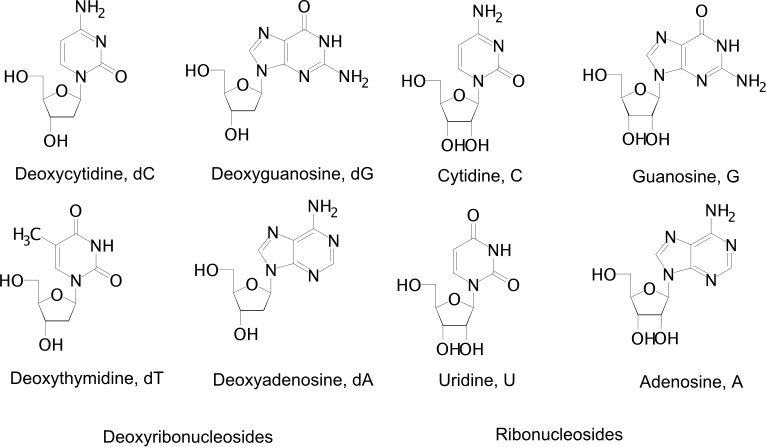
DNA consists of a phosphate backbone with different bases. Watson-Crick base pairs (meaning traditional base pairs) for DNA are dG:dC and dA:dT, where A, C, G, and T are adenine, cytosine, guanine, and thymine. In RNA, thymine is replaced by uracil to make up the traditional bases. Adenine and guanine are purines, meaning they have fused imidazole (5-member) and pyrimidine (6-member) rings. Cytosine, thymine, and uracil are pyridines with just the pyrimidine (6-member) ring. The different bases are shown below.
Figure: DNA Bases
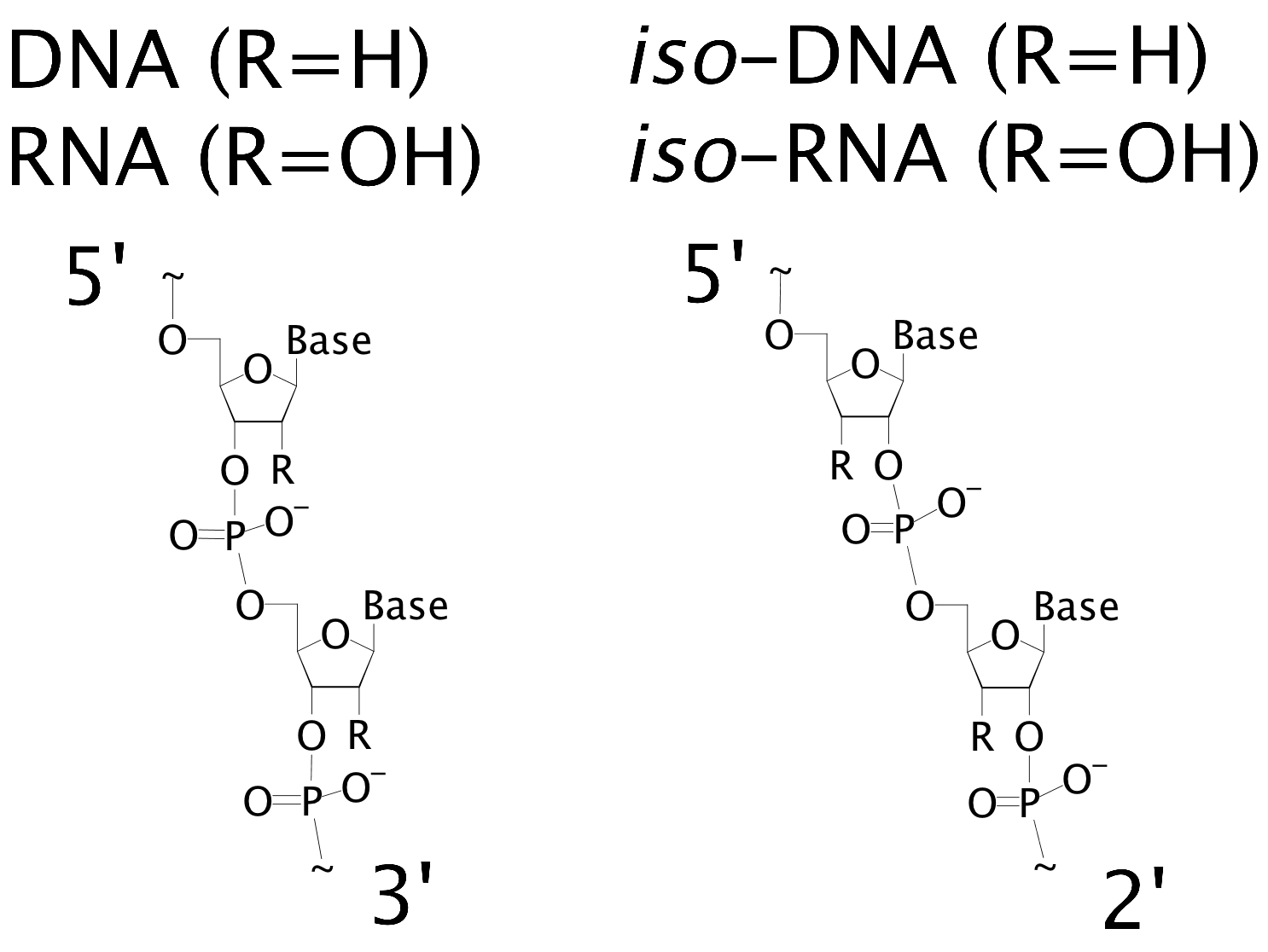
It is also important to know how the different DNA (and RNA) residues bind to each other. The typical backbone is linked through the 3’ and 5’ ends. In the iso forms, the backbone is linked through the 2’ and 5’ ends. These differences are shown in below.
Figure: DNA Binding
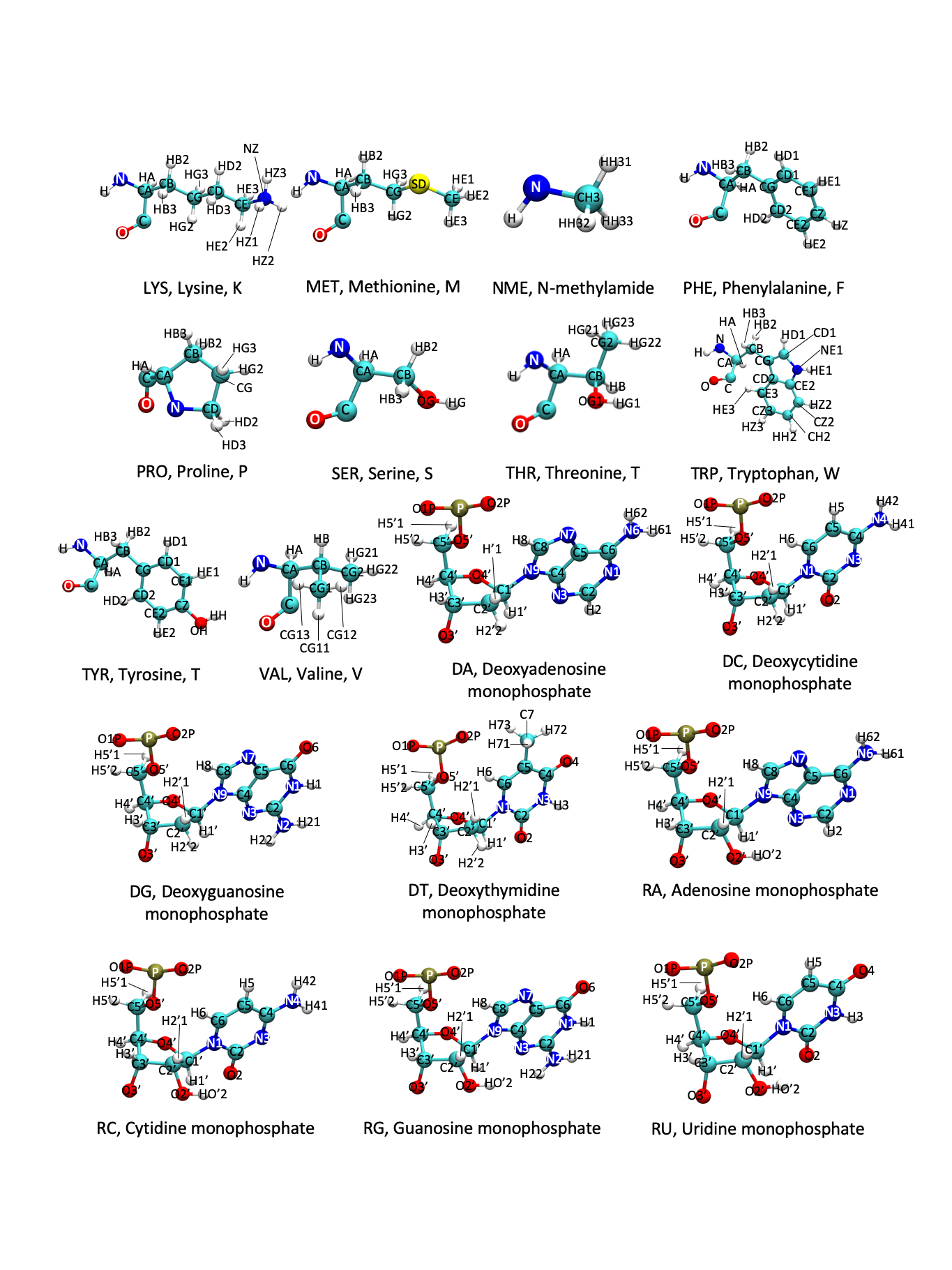
Amino acids are known by full names, single-letter codes, and three-letter codes. A list of these names and codes can be found with the skeletal structure in the figures at the end of the page.
The AMBER atom and residue naming is also shown in the following figures. Atom naming follows the Greek alphabet, starting with the alpha carbon (A) and moving on to beta (B), gamma (G), delta (D), epsilon (E), zeta (Z), and eta (H). In AMBER, there are additional 3-letter codes for residues with several protonation states that deviate from the traditional amino acid pattern. Additionally, the naming patterns for DNA and RNA bases are shown.
When a protein complexes with a prosthetic group, such as a DNA strand, then that structure is said to have apo and holo forms. In short, Mark came up with “Apo absent, holo has.”
Apoprotein: the protein part of an enzyme that is missing its prosthetic group. Think of it like a bear that’s missing their hat–they can exist without it, but they’re much happier with it.
Holoprotein: the apoprotein combined with its prosthetic group. The bear has located their hat again and is now wearing it.
Figure: Amino Acids
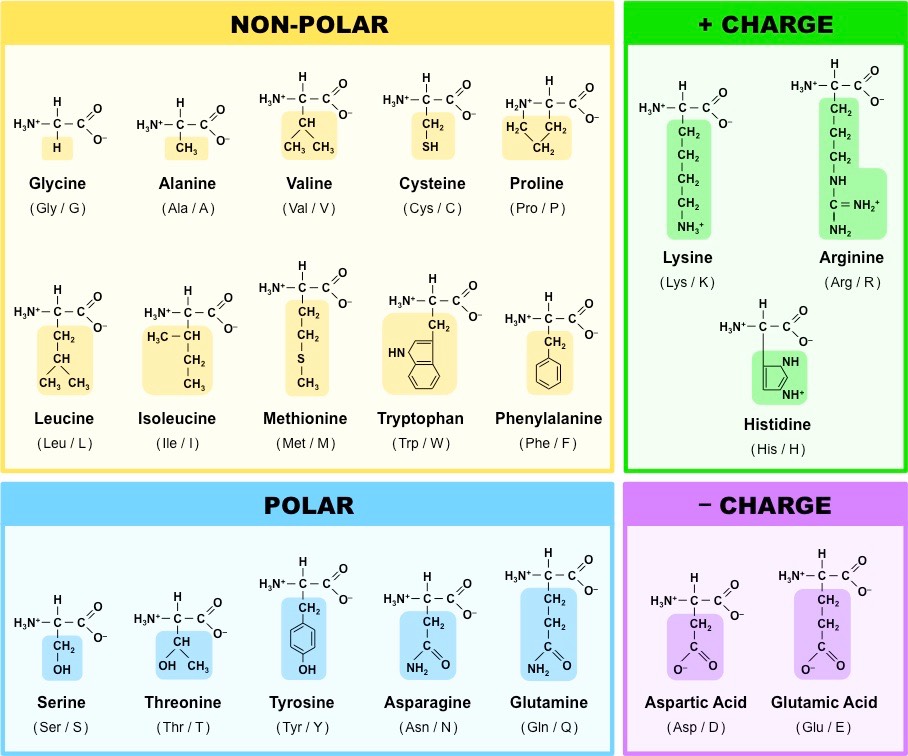
Figure: AMBER Amino Acids 1
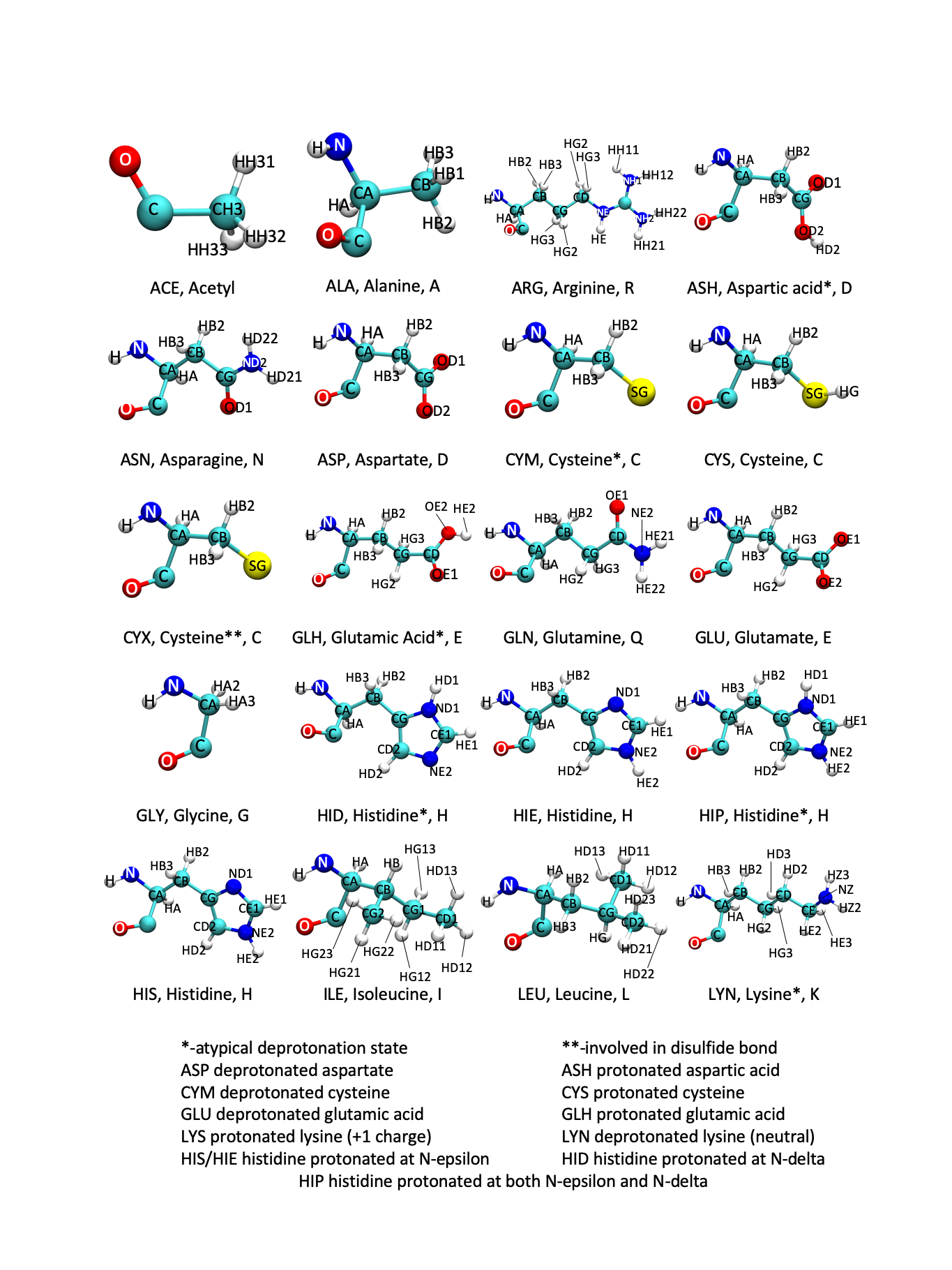
Figure: AMBER Amino Acids 2