Structure Preparation
Overview
Teaching: 120 min
Exercises: 0 minQuestions
How do I make a structure biologically relevant?
Do I need to add hydrogens?
Where do parameters come from?
How do I add ions and solvate a system?
Objectives
Modify a crystal structure to reflect realistic protonation states.
Check a structure for clashes.
Create parameters for a small, organic molecule.
Generate a solvated and charge-neutral structure with LEaP.
Highlights
- Where are the files?
- PDB Structure Background
- Initial Processing
- Possible Error
- What in the world are all those
sedcommands for? - Hold up, why did we remove
GOLagain? - PROPKA
- MolProbity
- LEaP
- “Decently described” sounds sketchy…
- Using
tleap - Testimonial from a Grad Student: Read the Error Messages
Where are the files?
Check out the web
PDB Structure Background
The ATOM and HETATM records in a PDB file contain the relevant information
for getting started with computational simulations.
ATOM 1 N HIS 4 30.098 2.954 7.707 1.00 0.00 N
The records start with ATOM or HETATM.
Next is the atom index or atom ID number.
Each atom index is unique to that atom in a simulation, but before the structure
is fully prepared, these numbers will often be non-unique.
Next is the atom name. Atom names should be unique within a given residue.
So, for a methyl group with 3 hydrogens, those hydrogens would likely be labeled
as H1, H2, and H3.
After the atom name comes the residue name.
AMBER uses unique residue names for residues in different protonation states,
but not every program or force field follows this method.
Next is the residue number.
These should also be unique once the structure is fully prepared.
The following three numbers are the X, Y, and Z coordinates for the atom.
The next two numbers are the occupancy and the temperature factor, respectively.
RCSB has a good explanation of these in their
PDB-101
site, but they are not important right now.
Finally, the element symbol for the atom is listed.
Initial Processing
Download 5y2s.pdb from RCSB.
RCSB has a dropdown for “Display Files” and “Download Files.”
Choose the PDB format.
PDB files have a lot of crystallographic information that can confuse MD
programs. The important lines start with ATOM, HETATM, TER, and END.
The clean_pdb.sh file is a script that includes the following command to
extract these lines from the PDB file and save them in a new file.
grep -e '^ATOM\|^HETATM\|^TER\|^END' $thing > $thing_clean
To run the clean_pdb.sh script, use:
bash clean_pdb.sh
or
./clean_pdb.sh
Possible Error
If you try the second option (./clean_pdb.sh), you may see:
-bash: ./clean_pdb.sh: Permission deniedWhen scripts are initially written, they do not have the appropriate permissions to be run. This is actually an anti-malware strategy! You don’t just want commands to be able to run all willy nilly. That’s why you then need to use another command,
chmodto modify the file permissions and allow it to be executed (run).chmod u+x clean_pdb.sh
Running the clean_pdb.sh script creates the 5Y2S_clean.pdb file.
What in the world are all those
sedcommands for?Each of those
sedcommands is for in-place editing of the file. The first block removes “B chain” residues, the second block renames the “A chain” residues as the normal residue name, and the third block renamesHOHtoWATand removes theGOL, which is an inhibitor. PDBs often have “A chain” and “B chain” residues, which offer alternative occupancies for an atom. These occupancies are kind of like saying 65% of the day someone sits at a desk, but the other 35% of the day they lay in their bed. You would likely care most about the time spent at the desk, but there are also times that knowing they’re in bed is important. For the purpose of this tutorial, we’re taking all the A-chain residues and ignoring all the B-chain residues. Every system is different, and every structure is different, so it’s better to make a case-by-case decision on which of the chains to use for a residue.Hold up, why did we remove
GOLagain?There are a few common inhibitors for enzymes, and
GOL(glycerol) is often one of them. You can check for potential inhibitors by reading the paper published with the crystal structure (if one exists…). This is one case where reading the methods section is critically important.
PROPKA
The crystal structure doesn’t come with hydrogens. This is because they both rotate frequently, and they are very, very small compared to everything else, so they’re not actually captured. Because of that, hydrogens are placed based on an educated guess, but protonation states are pH dependent. Thus, we need to use something to predict the proper protonation states for each titratable residue. Here, we’ll use PROPKA.
Go to the PROPKA webserver and select PDB2PQR.
Upload the 5Y2S_clean.pdb file.
These options should be used with the webserver:
- pH 7.0
- Use PROPKA to assign protonation states at provided pH
- Forcefield: AMBER
- Choose output: AMBER
- Deselect Remove the waters from the output file
Once the job has completed, there are a number of files to download.
The relevant output files have the pqr, stdout.txt and stderr.txt
extensions.
The pqr file contains the adjusted residue names.
Not to skip ahead in the tutorial (that’s why these files are provided 😃),
but you can use MDAnalysis to convert the output PQR back to a PDB.
This is what the propka-reintegration.py script looks like.
import MDAnalysis as mda
propka_output="../2-propka_output/p09m0bn8cm.pqr"
pdb_out="5Y2S_ph7_protonated.pdb"
system = mda.Universe(propka_output, format="PQR", dt=1.0, in_memory=True)
## Remove the segment IDs (w/o this, `SYST` gets annoyingly printed by element)
for atom in system.atoms:
atom.segment.segid = ''
system.atoms.write(pdb_out)
A physical inspection of the .pqr file from PROPKA reveals these REMARK lines:
REMARK 5 WARNING: PDB2PQR was unable to assign charges
REMARK 5 to the following atoms (omitted below):
REMARK 5 2157 ZN in ZN 301
REMARK 5 2158 C in CO2 302
REMARK 5 2159 O1 in CO2 302
REMARK 5 2160 O2 in CO2 302
REMARK 5 2161 C in CO2 303
REMARK 5 2162 O1 in CO2 303
REMARK 5 2163 O2 in CO2 303
This means that PROPKA couldn’t process the ZN or CO2 residues.
Because of that, they weren’t included in the PQR, and thus were not in the
converted PDB.
Make a copy of the converted PDB to add the ZN and CO2 residues into.
cp 5Y2S_ph7_protonated.pdb 5Y2S_ph7_nsa.pdb
Now, you can copy the ZN and CO2 atom lines from 5Y2S_clean.pdb into
5Y2S_ph7_nsa.pdb.
As part of this:
- Change the temperature factor (the number after XYZ coordinates) to
0.00 - Remove the A segment ID
- Change
HETATMtoATOM
The 5Y2S_clean.pdb ZN line
HETATM 2157 ZN ZN A 301 14.442 0.173 15.262 1.00 3.61 ZN
becomes
ATOM 2157 ZN ZN 301 14.442 0.173 15.262 1.00 0.00 ZN
You can leave the atom ID (2157) as-is.
Further programs will renumber everything.
MolProbity
Upload the cleaned PDB file (5Y2S_ph7_nsa.pdb) with the corrected pH AMBER
types to MolProbity.
As part of the structure processing by MolProbity, all of the hydrogens are
removed.
These hydrogens must then be re-added with the “Add hydrogens” button.
Selecting this button brings up a new page with options.
Choose Asn/Gln/His flips as the method.
In this particular case, the 5Y2S structure comes from x-ray crystallography.
Thus, select Electron-cloud x-H for the bond length.
Then you can hit Start adding H >.
MolProbity found evidence to flip several residues.
Select the ones you’d like to flip with the check-boxes (all recommended)
before hitting Regenerate H, applying only selected flips >.
Because of the addition of hydrogens and flips, a pop-up appears.
Select yes to download the new structure now.
LEaP
LEaP works best with a PDB that’s as close to true form as possible. The new structure downloaded from MolProbity has some issues with it. We need to rectify these things:
TERlines should be between the protein and any metals, ions, or waters- Remove the
flipandnewmarks that MolProbity prints - Remove
USERandREMARKfrom PROPKA and MolProbity - Use atom names that are consistent with what LEAP expects
- In this case, change
OWforWATtoO
- In this case, change
This PDB file was cleaned up through by deleting the USER and REMARK heading
lines, as well as performing these string replacements with vi.
:%s/ flip//g
:%s/ new//g
:%s/ OW WAT/ O WAT/g
The other important thing we need to consider about this structure is its active site. The CO2 residue is not native to AMBER, so we need to get parameters for it. If this were a residue that had hydrogens (like a drug), then the anticipated hydrogens would need to be added before it was parametrized. Additionally, the ZN residue in the active site is one that would benefit from additional parameters. In this case, the ZN if 4-coordinate with 2 HID, 1 HIE, and 1 WAT. This is adequately described by ZAFF (which has its own tutorial).
Generating CO2 Parameters
Since CO2 is a small, organic molecule, it should be decently described by GAFF.
“Decently described” sounds sketchy…
Good! You picked up on that! It can be sketch. GAFF stands for General AMBER Force Field. That means it’s generalized for organic compounds that contain C, N, O, H, S, P, F, Cl, Br and I. So, if you have something that’s tricky, or that you’d want more tailored parameters for, you should use RESP fitting to compute them. One way to achieve this is to use pyRED.
To get the GAFF parameters, run antechamber.
Each of the flags gives the program information it needs to run.
| Flag | Purpose |
|---|---|
| -i | input file |
| -fi | file type of input file |
| -o | output file |
| -fo | file type of output file |
| -c | calculation type |
| -s | verbosity |
| -nc | net charge |
| -m | multiplicity |
antechamber -i CO2.pdb -fi pdb -o CO2.mol2 -fo mol2 -c bcc -s 2 -nc 0 -m 1
Ordinarily, after running antechamber, you run parmchk to make sure that
all of the necessary force field descriptors are present.
parmchk -i CO2.mol2 -f mol2 -o CO2.frcmod
This file contains no information, as everything required is covered by GAFF.
Incorporate ZAFF
ZAFF is the Zinc AMBER Force Field (read more). There are several parameter sets for for different 4-coordinate zinc complexes.
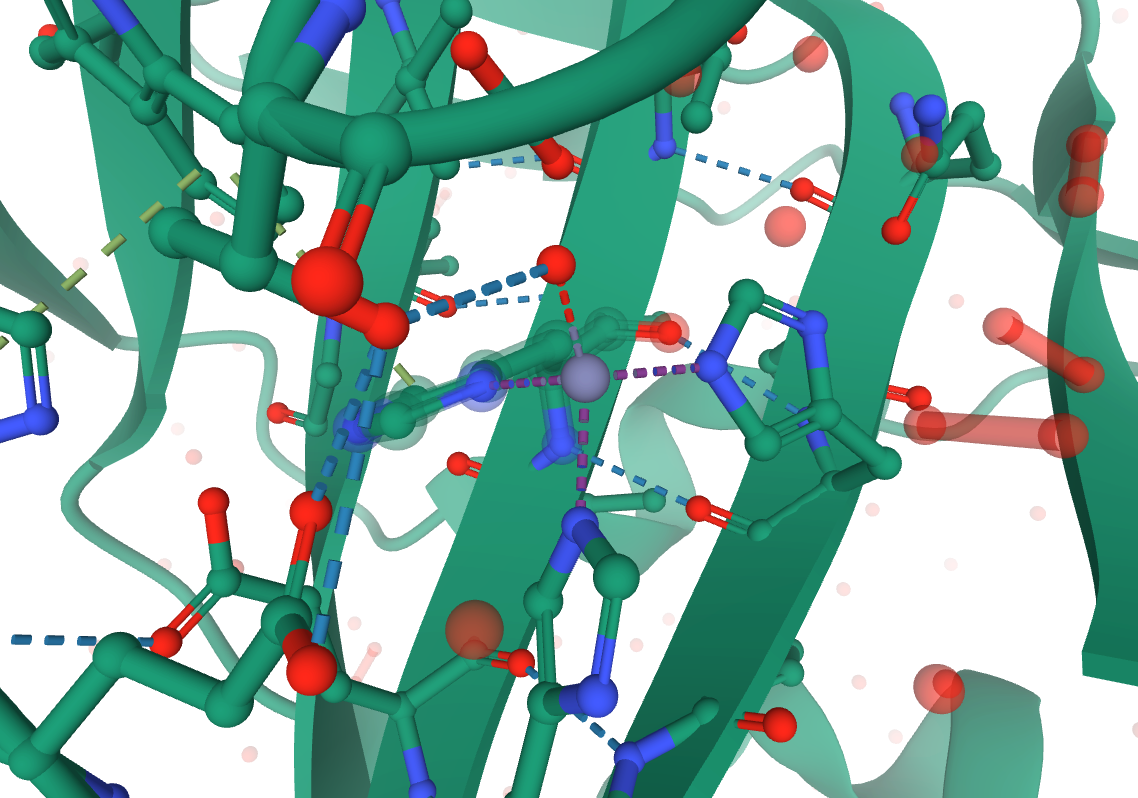
Looking at the 3D view on the RCSB website, you can identify that HID 94, HID 96, HIE 119, and WAT 455 are all coordinated to ZN 301 in the original structure. The 3 histidine, 1 water coordination is best described by ZAFF Center ID 4.
Image citation: 5Y2S, doi: 10.2312/molva.20181103, RCSB PDB.
Since PROPKA and MolProbity maintained the residue numbering, we can use those RESIDs to rename the coordinated residues as needed for ZAFF. Only the residue name (for the entire residue) needs to change.
| ResNum | Original ResName | New ResName |
|---|---|---|
| 94 | HID | HD4 |
| 96 | HID | HD5 |
| 119 | HIE | HE2 |
| 455 | WAT | WT1 |
| 301 | ZN | ZN6 |
Before moving on with LEaP, we need to download the parameter files for ZAFF. These can be downloaded from the ZAFF tutorial as a prep and frcmod. They are also included in the files downloaded for this lesson.
Using tleap
tleap is is the terminal form of LEaP.
There is also xleap, which uses a GUI.
Most of the time, people use an input script with tleap, which is evoked like:
tleap -f tleap.in
The tleap.in file for used here looks like:
source leaprc.gaff
source leaprc.protein.ff14SB
addAtomTypes { { "ZN" "Zn" "sp3" } { "N5" "N" "sp3" } { "N6" "N" "sp3" } { "N7" "N" "sp3" } }
source leaprc.water.tip3p
loadamberprep ZAFF.prep
loadamberparams ZAFF.frcmod
CO2 = loadmol2 CO2.mol2
c = loadpdb 5Y2S_ph7_mp_ZAFF.pdb
bond c.261.ZN c.94.NE2 ## HID NE2
bond c.261.ZN c.96.NE2 ## HID NE2
bond c.261.ZN c.119.ND2 ## HIE ND2
bond c.261.ZN c.318.O ## WAT O
savepdb c 5Y2S_vac.pdb
saveamberparm c 5Y2S_vac.prmtop 5Y2S_vac.inpcrd
addions c CL 0.0
solvatebox c TIP3PBOX 12.0
savepdb c 5Y2S_wat.pdb
saveamberparm c 5Y2S_wat.prmtop 5Y2S_wat_init0.rst
quit
First, the relevant force field information is sourced, and any new atom types
are defined.
Next, the information for ZAFF and CO2 are loaded in.
The CO2 = part corresponds to the 3-letter code that is in the PDB for that
residue.
Next, the protein is loaded in using a variable name, c.
Any variable name is fine, but if you update it, it’d need to be updated in
all of the remaining lines.
The bond lines generates a bond between the zinc and each of the atoms it
is coordinated to, which is an aspect of the ZAFF model.
The save lines save structures in various states.
There’s a first save before the ions and water are added, known as a “dry” or
“vacuum” structure.
Then, those ions (where 0.0 says to neutralize the charge) and waters are
added (where 12.0 specifies that water should extend 12.0 angstroms from the
protein’s surface).
These additions are saved in a “wet” or “solvated” structure.
LEaP does accept comments in these files, using #.
If you run this script on the provided files, it should work. However, if you were doing this on your own, you would likely encounter this error:
WARNING: The unperturbed charge of the unit: 0.998000 is not zero.
FATAL: Atom .R<NHIE 4>.A<HD1 20> does not have a type.
Failed to generate parameters
Parameter file was not saved.
Quit
The way to solve this error is to delete the HD1 hydrogen of the very first
residue, HIE 4, so that it can be rebuilt.
This is an error that can sometimes occur with first and last protein residues.
Testimonial from a Grad Student: Read the Error Messages
97.9% of my problems would be solved if I just read the error messages. Often, they tell you exactly what you need to do to fix it. If they don’t, I strongly recommend Google. Google is your best friend. Well, maybe not your best friend. I don’t even know you. I guess what I’m saying is that you’re probably not the first person to have an error of a specific type, which is why someone programmed an error message. Between the message, the internet, and the source code, there’s a lot of “clues” out there as to what’s going wrong.
Key Points
PROPKA is one way to calculate protonation states.
MolProbity can be used to check a structure for clashes, and make relevant ring flips.
Non-standard residues must have relevant hydrogens before parametrization.
For the final structure, LEAP will add hydrogens (and other missing sidechain atoms) based on residue definitions.